Project: Autoregressive Hidden Markov Model implementation
Repository: RoLi lab
Freely behaving animals don't just move randomly—they transition through distinct behavioral states organized at multiple temporal scales. A zebrafish might cycle between sleep and wake over hours, but within those periods, it's constantly switching between exploration and exploitation, movement and rest, active swimming and quiescence. Each of these states has characteristic behavioral signatures, but how do we identify them from the continuous stream of movement data?
This project tackles that challenge by developing and applying autoregressive hidden Markov models (ARHMMs) to identify behavioral motifs in freely swimming larval zebrafish. Unlike classic HMMs, ARHMMs capture the fact that what happens next in behavior often depends on what just happened—a crucial feature for understanding behavioral sequences. We implemented this method in Julia and applied it to high-resolution behavioral tracking data to uncover the hidden states that organize zebrafish behavior.
The Challenge: From Continuous Signals to Discrete States
When you watch a zebrafish swim freely in an arena, you're seeing a complex, continuous stream of behavior. The fish moves through space, its tail undulates, its eyes track or saccade, and its body posture changes constantly. But underlying this continuous behavior are discrete behavioral states—periods of active exploration, quiescent rest, directed swimming, and more. The challenge is identifying where one state ends and another begins.
Traditional approaches often rely on simple thresholds (e.g., "if speed > X, then active"). But behavior is more nuanced than that. A fish might be moving slowly but actively exploring, or moving quickly but in a stereotyped pattern. We needed a method that could capture the multidimensional nature of behavior and identify states based on patterns across multiple behavioral features simultaneously.
Why Autoregressive HMMs? Capturing Behavioral Dependencies
Hidden Markov Models are powerful tools for identifying hidden states from observed sequences. In a classic HMM, the probability of observing a particular behavior at time t depends only on the current hidden state. But behavior doesn't work that way—what you do next often depends on what you just did. A fish that just made a saccade is more likely to continue moving; a fish that just stopped is more likely to remain quiescent.
That's where autoregressive HMMs (ARHMMs) come in. In an ARHMM, the probability of an observation depends on both the current hidden state and the previous observation. This allows the model to capture sequential dependencies—the fact that behaviors have momentum, that transitions between states are structured, and that the context of what just happened matters for what comes next.
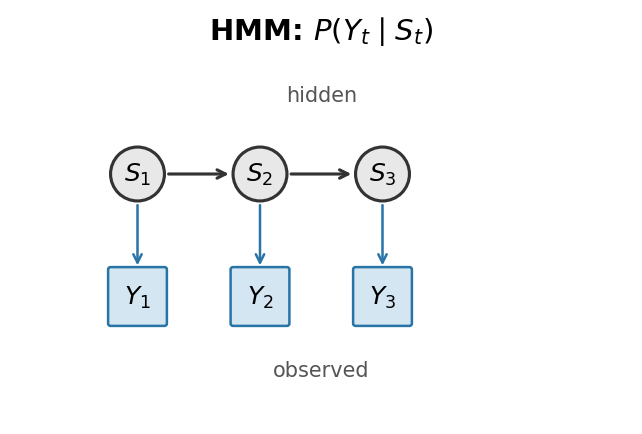
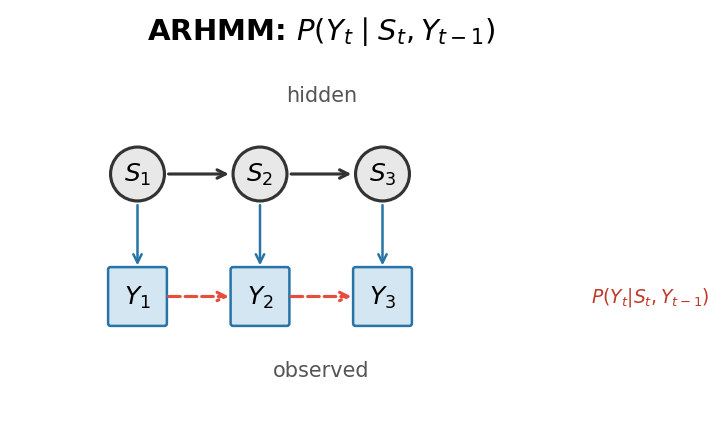
We implemented both HMM and ARHMM variants in Julia, with full parameter estimation (Baum–Welch algorithm), state decoding (Viterbi algorithm), and tools for discretizing continuous behavioral features using Gaussian mixture models. The package is available on GitHub and provides a complete pipeline from raw behavioral data to inferred state sequences.
Recording Behavior at High Resolution
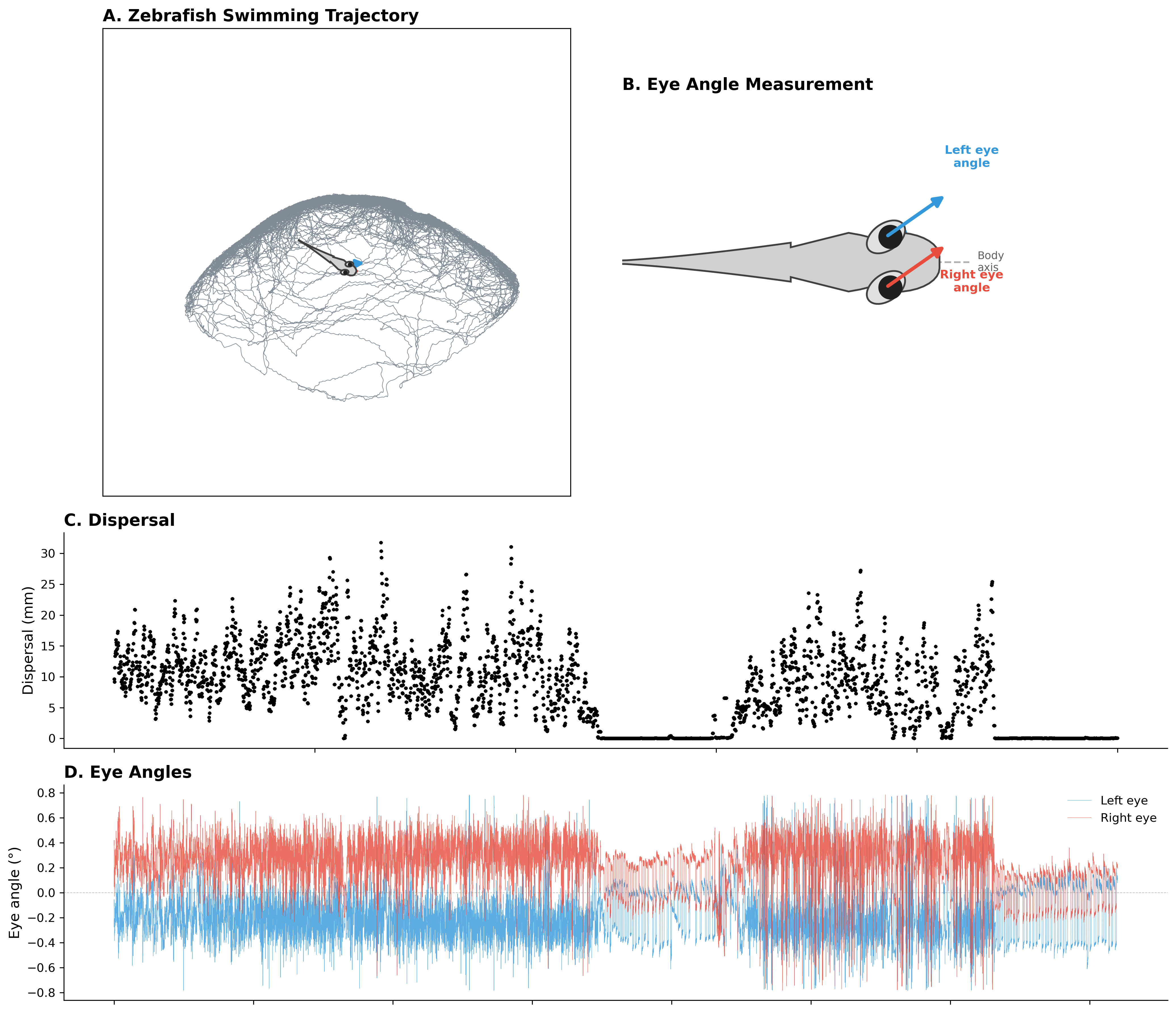
To apply this method, we recorded freely swimming larval zebrafish in a featureless arena (30 mm × 30 mm) at high spatiotemporal resolution. We tracked multiple behavioral parameters simultaneously:
- Speed and dispersal: How much the animal moves within a time window, capturing overall activity level
- Tail and heading: Body orientation and movement direction, revealing directed vs. undirected movement
- Eye movement: Left and right eye angles, allowing us to detect saccades and conjugate eye movements
From these raw signals, we derived three key observables for the ARHMM: dispersal (movement magnitude), heading velocity (smoothed directional movement), and saccade rate (frequency of conjugate saccades in the eye-angle traces). These three features capture different aspects of behavior—overall activity, directed movement, and eye movement patterns—that together define distinct behavioral states.
From Continuous to Discrete: Building the Model
The first step was converting our continuous behavioral signals into discrete symbols that the ARHMM could work with. We used a Gaussian mixture model (GMM) to cluster the multidimensional behavioral space into discrete categories. Each time point gets assigned to one of these categories based on its combination of dispersal, heading velocity, and saccade rate.
Then we fit a 5-state ARHMM to this discrete sequence. The model learns:
- State transition probabilities: How likely is the fish to switch from one behavioral state to another?
- Emission probabilities: Given a hidden state and the previous observation, what's the probability of observing each behavioral symbol?
- Initial state distribution: What states does the fish start in?
Using the Viterbi algorithm, we then decode the most likely sequence of hidden states given the observed behavioral data. This gives us a state assignment for every time point, revealing when the fish transitions between different behavioral motifs.
What We Found: Organized Behavioral Motifs
The ARHMM successfully identified extended bouts of distinct behavioral activity with smooth transitions between them. Rather than random switching, the model revealed that zebrafish behavior is organized into coherent, state-like motifs that persist over time and transition in structured ways.
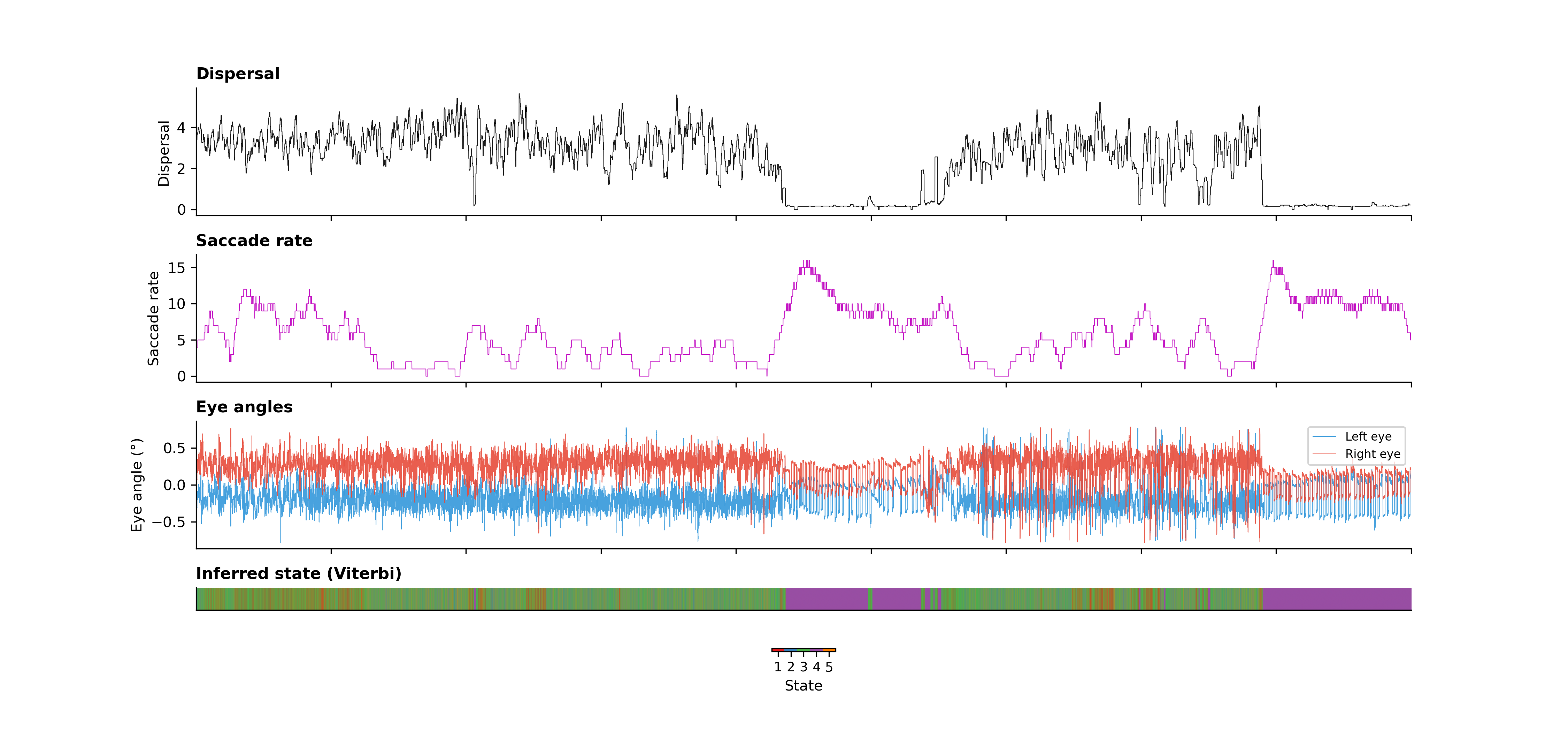
Each of the five inferred states captures a different behavioral pattern. Some states correspond to active exploration—high dispersal, frequent saccades, directed movement. Others correspond to quiescent periods—low dispersal, minimal movement, stable eye positions. The transitions between states aren't random; they follow patterns that reflect the organization of behavior at multiple temporal scales.
This approach provides a powerful framework for understanding how behavior is organized. By identifying the hidden states that structure behavioral sequences, we can ask questions about how these states are regulated, how they change across conditions, and how they relate to underlying neural activity. The ARHMM doesn't just describe behavior—it reveals the structure that organizes it.
Implementation and Tools
The complete implementation is available as a Julia package on GitHub. The package provides:
- HMM and ARHMM models: Full implementations with parameter estimation and state decoding
- Baum–Welch algorithm: Expectation-maximization for learning model parameters from data
- Viterbi algorithm: Finding the most likely state sequence
- GMM discretization: Converting continuous behavioral features into discrete symbols
- Model selection: Log-likelihood and AIC for comparing models
The package is designed to be flexible and extensible, making it easy to apply ARHMMs to other behavioral datasets or adapt the method for different types of sequential data. All code is documented and includes example notebooks showing how to apply the method to zebrafish behavioral data.
Why This Matters: Understanding Behavioral Organization
Behavior isn't just a continuous stream—it's organized into discrete, coherent states that persist over time and transition in structured ways. By identifying these states using ARHMMs, we can understand how behavior is organized at multiple temporal scales, from rapid transitions between movement and rest to longer-term cycles of exploration and exploitation.
This approach has broad applications. It can be used to identify behavioral states in any animal, to compare behavior across conditions or genotypes, and to link behavioral states to underlying neural activity. The method provides a quantitative, data-driven way to parse behavior into interpretable motifs, opening up new questions about how behavior is generated, regulated, and organized.
For zebrafish specifically, understanding these behavioral motifs helps us understand how sleep and wake states are organized, how animals explore their environment, and how behavioral states relate to neural activity patterns. By combining this behavioral analysis with whole-brain imaging, we can begin to map the neural circuits that generate and regulate these behavioral states.